Overview
In this lesson, I introduce the independent samples t test, which you use to test for a difference in the means of two independent samples. You will generate fake data to practice using the t test.
The Problem
You predict that people who feel powerless will feel more sympathy for someone else who is upset than people who feel powerful.
To test this prediction, you run an experiment with two conditions. In the powerless condition (n = 60), subjects write about a time when they were subordinate to someone else. In the powerful condition (n = 60), subjects write about a time when they had power over someone else.
After the power manipulation, subjects read a letter from someone else who says that he is upset because he recently lost his job. You ask subjects to report how much they feel sympathetic, compassionate, and empathetic on a five-point Likert-type scale (0=not at all, 4=extremely).
Your hypothesis is that subjects in the powerless condition will feel more sympathetic than those in the powerful condition on average. Your null hypothesis is that subjects in the two conditions will feel equally sympathetic on average.
Step 1: Generate the Fake Data
First, let’s create the grouping variables. We have 60 subjects per condition, so we need to create a variable that has 60 observations in the powerless condition and 60 observations in the powerful condition.
Remember that you can make different kinds of objects in R. When you’re coding groups, you have a choice to make: should you make the grouping variable a numeric object or a factor? There are some conditions where you will want your group variable to be numeric, but for this simple problem it’s probably easier to code it as a factor so that you can see the group labels. The good news is that when you’re using R it’s easy to convert a factor into a numeric object later if needed.
There are a couple of ways that you can create a factor, so just use whatever makes the most sense to you.
# Option 1: Create a string variable and then convert it into a factor
> groups <- c(rep('powerless',60), rep('powerful',60))
> groups <- factor(groups)
If you look at the help file for the rep() function, you’ll see that two arguments are rep(x, times), meaning that it will replicate whatever you put into the first argument (‘powerless’ or ‘powerful’) the number of times that you enter into the second argument (60 times). Alternatively, you can use rep(x, each) to replicate each element of the first argument 60 times, as in the following:
> groups <- rep(c('powerless','powerful'), each=60)
> groups <- factor(groups)
# Option 2: Create the factor directly
> groups <- factor(rep(c('powerless','powerful'), each=60))
Second, let’s create the dependent variables. We need to create responses to the three emotions: sympathetic, compassionate, empathetic. We’ll start by using the rnorm(n,mean,sd) function to randomly sample data from normal distributions. The first argument n sets the sample size. The second argument mean sets the mean of the population that we’ll sample from. The third argument sd sets the standard deviation of the population that we’ll sample from. Our first 60 observations will be for the powerless group and the second 60 observations will be for the powerful group.
Before we generate the dependent variable data, we’ll set the seed to a specific number. This number will affect the way that your computer generates random data. In particular, if you set the seed to the number that I specify here, you should get the same “random” results that I post here.
> set.seed(1111)
> sympathetic <- c(rnorm(60,3,.5), rnorm(60,2.5,.5)) # the powerless group has mean=3 and sd=.5; the powerful group has mean=2.5 and sd=.5
> compassionate <- c(rnorm(60,2.6,.5), rnorm(60,2.3,.5))
> empathetic <- c(rnorm(60,2.2,.5), rnorm(60,1.4,.5))
If you look at your results, you should notice a couple of problems. We said that subjects completed a 0-4 Likert-type scale, but some of the scores are greater than 4 and all of the observations are decimals rather than integers. We’ll have to fix this before we move on. Each time, we’ll make new variables for sympathetic, compassionate, and empathetic by applying some function to the existing variables.
First, make it so that if an observation is greater than 4, it is set equal to 4. We’ll use the ifelse(test, yes, no) function. The first argument is a testing condition that can be TRUE or FALSE. The second argument specifies what happens if the testing condition is TRUE. The third argument specifies what happens if the testing condition is FALSE.
> sympathetic <- ifelse(sympathetic > 4, 4, sympathetic) # if the value of an element of the variable sympathetic is greater than 4, then set it equal to 4; otherwise, set it equal to its original value
> compassionate <- ifelse(compassionate > 4, 4, compassionate)
> empathetic <- ifelse(empathetic > 4, 4, empathetic)
Next, we’ll use the round(x, digits) function to round the decimals to integers.
> sympathetic <- round(sympathetic, digits=0)
> compassionate <- round(compassionate, digits=0)
> empathetic <- round(empathetic, digits=0)
Finally, let’s combine the grouping variable and the dependent variables into a single data frame. We’ll also clean up our workspace by using the rm() function to get rid of the original variables.
> power.data <- data.frame(groups, sympathetic, compassionate, empathetic)
> rm(groups, sympathetic, compassionate, empathetic)
From now on if you want to access the variables, you’ll have to pull them from the power.data data frame.
> sympathetic # just entering the variable name returns "Error: object 'sympathetic' not found" because it is inside the power.data data frame
> power.data$sympathetic # this works because the dollar sign says "Look for sympathetic inside the object power.data"
> with(power.data, sympathetic) # this also works because the with() function says "Do everything inside the object specified by the first argument," which in this case is the data frame power.data
Now let’s say that you think the items sympathetic, compassionate, and empathetic are all measuring the same emotional reaction in slightly different ways. You’ll want to combine them by summing them or by finding the average of all three. It doesn’t really matter which, but to keep our overall sympathy measure on the same scale as the individual items (0-4), we’ll create a new variable in our data frame that has the mean of the three emotion items.
Your data are organized in the data frame as rows of participants and columns of variables. We want to combine the three emotion items across participants, so we want our new variable to show the average of the three items for each row (i.e., each participant). We can’t do this with the mean() function because it will give us the average of all three variables combined across all participants rather than the average for each participant separately. Instead, we’ll use the rowMeans() function.
> power.data$sympathy <- with(power.data, rowMeans(cbind(sympathetic, compassionate, empathetic))) # the cbind() function treats the variables as columns of data, which is required by the rowMeans() function; there is also an rbind() function that would treat them as rows of data
Step Two: Check Assumptions
The independent samples t test makes three assumptions
- Independence. All observations are independent of each other, meaning that the probability of an observation taking on a specific value does not depend on the value of any other observation.
- Normality. The data in each group are normally distributed.
- Equal variances. The data from the two groups are equally variable. The complicated term that you’ll sometimes see for this is “homoscedasticity.”
In order for the results of the t test to be meaningful, we need to check these assumptions.
Checking independence in this case has more to do with your research design than a statistical check. How might your data violate the independence assumption? One clear example is if the same person participates in your study twice. If that person has a tendency to be more sympathetic in general, then they are likely to be extra sympathetic both times that they participate. Those two observations of the person’s sympathy are linked and independence is violated.
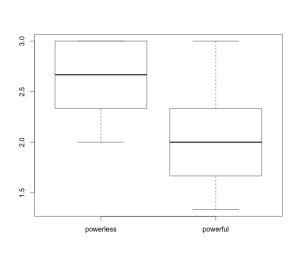
There are a few ways that you can check for normality and equal variances, but the easiest thing to do is to plot a boxplot of our composite sympathy score by group.
> boxplot(sympathy ~ group, data=power.data)
Recall that our normal distribution looks like this.
You can think of the boxplots as looking at the distribution of our data from above instead of from the side. Each box represents the middle 50% of the data for each group, which is called the interquartile range. The dark line is the median (the middle number for each group). The top and bottom whiskers reach out to the farthest points that are within 1.5 times the length of the interquartile range from the edges of the box. If you see any dots outside of the whiskers, then those are potential outliers (there are none in this case).
For the data in each group to be normally distributed, the boxplot for each group should look symmetric. This means that the medians (the dark lines) should be in the center of each box and the top and bottom whiskers within each group should be the same length. In our plot here the medians look okay, but the whiskers are a bit off. In the powerless group there is no top whisker because the box hits the maximum value in the data (which is 3). In the powerful group the top whisker is a bit longer than the bottom whisker, though the difference is not terrible. There might be a violation of the normality assumption in these data, but for the sake of this lesson we’ll pretend that things are okay.
For equal variances to hold, the interquartile ranges and whiskers should be the same length across the two groups. In our plot here, the interquartile ranges and the bottom whiskers look okay. However, the top whisker is longer in the powerful group than in the powerless group. There might be a violation of the equal variances assumption in these data. In practice, this will be okay. The classic t test assumes that variances are equal across groups, but there is also the Welch t test that does not assume equal variances. I’ve done some research showing that you tend to make better decisions if you just use the Welch t test at all times, so I’ll encourage you to use that t test exclusively.
Step Three: Run the t test
If our data are normally distributed and have equal variances across groups, then we can run our t test to compare the means of the two groups. First, we’ll look at the means and standard deviations of our groups using the by(x, group, function) function.
# Remember to use the with() function to access the variables that are inside the power.data data frame
> with(power.data, by(sympathy, groups, mean))
groups: powerless
[1] 2.622222
---------------------------------------------------
groups: powerful
[1] 2.077778
> with(power.data, by(sympathy, groups, sd))
groups: powerless
[1] 0.309709
---------------------------------------------------
groups: powerful
[1] 0.3801411
To run the classic t test in R, you use the t.test(y ~ x, var.equal=TRUE, data=myData) function.
> t.test(sympathy ~ groups, var.equal=TRUE, data=power.data)
Two Sample t-test
data: sympathy by groups
t = 8.6008, df = 118, p-value = 3.948e-14
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
0.4190897 0.6697992
sample estimates:
mean in group powerless mean in group powerful
2.622222 2.077778
In these data, the classic t test shows that there is a difference between the means of the two groups. The p value is less than the .05 cutoff and the 95% confidence interval does not contain 0 (more on confidence intervals in a future lesson). Now let’s run the Welch t test, which does not assume separate variances. All we need to do is change the value of the var.equal argument in to FALSE.
> t.test(sympathy ~ groups, var.equal=FALSE, data=power.data)
Welch Two Sample t-test
data: sympathy by groups
t = 8.6008, df = 113.37, p-value = 5.066e-14
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
0.4190366 0.6698522
sample estimates:
mean in group powerless mean in group powerful
2.622222 2.077778
What changed? The Welch t test adjusts the degrees of freedom to the extent that the variances of the two groups are unequal, and the degrees of freedom can have a decimal now. Additionally, the Welch t test uses a different standard error. As a consequence, the p value and confidence interval are slightly different, even though the t value did not change. In this case, the Welch t test also shows that there is a difference between the two groups. In practice, the two tests will almost always lead to the same decision when either the sample sizes or variances are equal, but when they lead to different decisions it is always safer to go with the Welch t test.
Step Four: Write it Up
Here is a sample of how I would write the results (you have my permission to use this format in your own writing if you would like). Using the Welch t test is not standard, even though it’s the better decision, so you need to specify that you used it in a paper.
To test whether there was a difference in sympathy between subjects in the powerless condition and those in the powerful condition, I ran a separate variances t test. Subjects in the powerless condition felt more sympathy (M = 2.62, SD = .31) than subjects in the powerful condition (M = 2.08, SD = .38, t(113.37) = 8.60, p < .001, 95% CI [.42, .67]).